If you choose the UMAP curve parameters to be a = 1 and
b = 1, you get back the Cauchy distribution used in t-Distributed Stochastic
Neighbor Embedding and LargeVis. This also happens
to significantly simplify the gradient leading to a noticeable
speed-up.
For MNIST:
library(uwot)
# install snedata package from github
# pak::pkg_install("jlmelville/snedata")
mnist <- snedata::download_mnist()I saw the optimization time drop from 66 seconds with UMAP:
mnist_umap <- umap(mnist, n_neighbors = 15)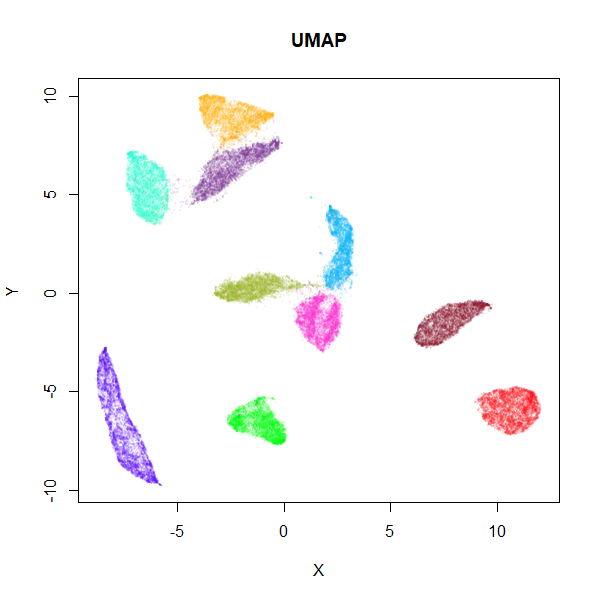
MNIST UMAP
to 18 seconds with t-UMAP:
mnist_tumap <- tumap(mnist, n_neighbors = 15)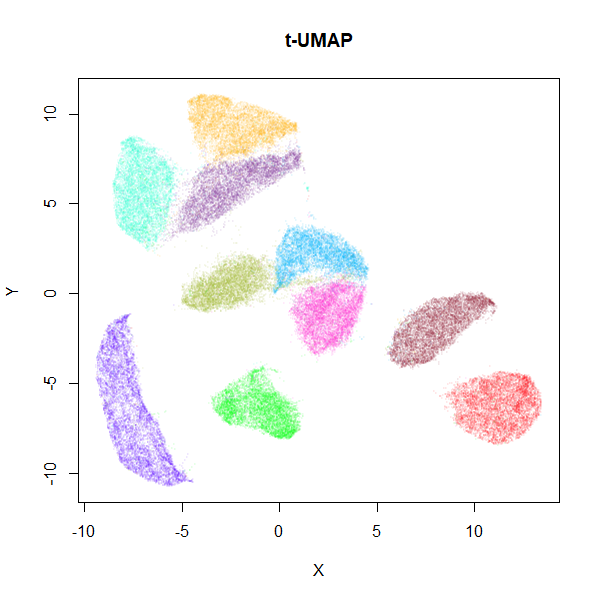
MNIST t-UMAP
You will still spend most of the time in the nearest neighbor search,
so you will really see a difference in terms of total time with larger
values of n_epochs. The trade off, as you can see, is that
you will see larger, more spread-out clusters than with the typical UMAP
settings (they’re still more compact than you see in t-SNE, however). I
think it’s worth the trade-off.
Note that using umap(a = 1, b = 1) doesn’t use the
simplified gradient, so you won’t see any speed-up that way.
Some examples comparing UMAP and t-UMAP are in the examples article.