An R implementation of the Uniform Manifold Approximation and Projection (UMAP) method for dimensionality reduction of McInnes et al. (2018). Also included are the supervised and metric (out-of-sample) learning extensions to the basic method. Translated from the Python implementation.
News
November 10 2025 uwot version 0.2.4 has been released to CRAN. This was mainly to avoid a test potentially starting to fail due to an incorrect use of testthat. However, as part of some other small fixes, optional dependencies (e.g. RSpectra) will now be correctly detected and used if installed even if not loaded. This could have an effect on output (but the old behavior was a bug).
February 24 2025 uwot version 0.2.3 has been released to CRAN. This release mainly fixes some bugs, including one that was causing an error with an upcoming version of R-devel. One new feature: set rng_type = "deterministic" to use a deterministic sampling of vertices during the optimization phase which will give faster and more reproducible output at the cost of accuracy. The idea for this came straight from Leland McInnes via Reddit.
Installing
From CRAN
install.packages("uwot")From github
uwot makes use of C++ code which must be compiled. You may have to carry out a few extra steps before being able to build this package:
Windows: install Rtools and ensure C:\Rtools\bin is on your path.
Mac OS X: using a custom ~/.R/Makevars may cause linking errors. This sort of thing is a potential problem on all platforms but seems to bite Mac owners more. The R for Mac OS X FAQ may be helpful here to work out what you can get away with. To be on the safe side, I would advise building uwot without a custom Makevars.
install.packages("devtools")
devtools::install_github("jlmelville/uwot")Example
library(uwot)
# umap2 is a version of the umap() function with better defaults
iris_umap <- umap2(iris)
# but you can still use the umap function (which most of the existing
# documentation does)
iris_umap <- umap(iris)
# Load mnist from somewhere, e.g.
# devtools::install_github("jlmelville/snedata")
# mnist <- snedata::download_mnist()
mnist_umap <- umap(mnist, n_neighbors = 15, min_dist = 0.001, verbose = TRUE)
plot(
mnist_umap,
cex = 0.1,
col = grDevices::rainbow(n = length(levels(mnist$Label)))[as.integer(mnist$Label)] |>
grDevices::adjustcolor(alpha.f = 0.1),
main = "R uwot::umap",
xlab = "",
ylab = ""
)
# I recommend the following optional packages
# for faster or more flexible nearest neighbor search:
install.packages(c("RcppHNSW", "rnndescent"))
library(RcppHNSW)
library(rnndescent)
# Installing RcppHNSW will allow the use of the usually faster HNSW method:
mnist_umap_hnsw <- umap(mnist, n_neighbors = 15, min_dist = 0.001,
nn_method = "hnsw")
# nndescent is also available
mnist_umap_nnd <- umap(mnist, n_neighbors = 15, min_dist = 0.001,
nn_method = "nndescent")
# umap2 will choose HNSW by default if available
mnist_umap2 <- umap2(mnist)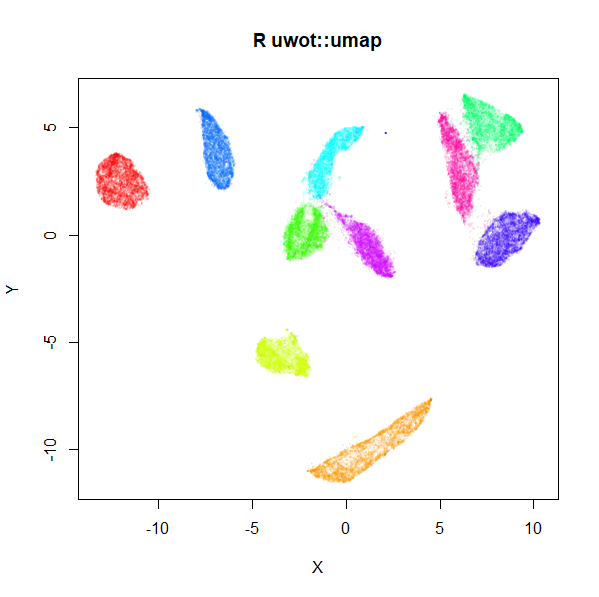
Documentation
https://jlmelville.github.io/uwot/. For more examples see the get started doc. There are plenty of articles describing various aspects of the package.
Citation
If you want to cite the use of uwot, then use the output of running citation("uwot") (you can do this with any R package).
See Also
- The UMAP reference implementation and publication.
- The UMAP R package (see also its github repo), predates
uwot’s arrival on CRAN. - Another R package is umapr, but it is no longer being maintained.
- umappp is a full C++ implementation, and yaumap provides an R wrapper. The batch implementation in umappp are the basis for uwot’s attempt at the same.
-
uwotuses the RcppProgress package to show a text-based progress bar whenverbose = TRUE.