Using HNSW for nearest neighbors
It’s a (Hierarchical Navigable) Small World After All
Source:vignettes/articles/hnsw-umap.Rmd
hnsw-umap.RmdHNSW for nearest neighbors
The Hierachical Navigable Small World (HNSW) method for finding approximate nearest neighbors is a popular due to its speed and good performance. A header-only C++ implementation by the HNSW authors can be found at hnswlib, and R-bindings are available in the package RcppHNSW.
If you would like to use HNSW for nearest neighbor search,
uwot now integrates with RcppHNSW as an
optional dependency. You just need to install the RcppHNSW
package:
install.packages("RcppHNSW")uwot will now be able to use it if you set
nn_method = "hnsw", e.g.
library(uwot)
# doesn't use HNSW
iris_umap <- umap(iris)
# does use HNSW
iris_umap_hnsw <- umap(iris, nn_method = "hnsw")MNIST example
For a more involved example we’ll use the MNIST digits data set for
this article, for which you will need to install the
snedata package from GitHub:
# install.packages("pak")
pak::pkg_install("jlmelville/snedata")
# or
# install.packages("devtools")
# devtools::install_github("jlmelville/snedata")Then, download the MNIST dataset (this requires access to the internet), and we will split the data into the traditional training/test split used with this dataset.
mnist <- snedata::download_mnist()
mnist_train <- head(mnist, 60000)
mnist_test <- tail(mnist, 10000)mnist is a dataframe where the first 768 columns contain
the pixel data for each image, and the last column is a factor
Label, which contains the digit the image represents.
UMAP on training data with HNSW
As noted above, you only need to use nn_method = "hnsw"
to use HNSW. But I am going to also set some other non-default
parameters:
-
verbose = TRUE– I just like to know what’s going on. -
n_sgd_threads = 6is the number of threads to use in the optimization step. -
batch = TRUEis an alternative optimization method that gives a bit more repeatable results when settingn_sgd_threadsto greater than one. -
n_epochs = 500is the number of epochs to use in the optimization step. I find that whenbatch = TRUE, you need to use a larger number of epochs than the usual default (which for MNIST-sized datasets would ben_epochs = 200). -
ret_model = TRUEis a parameter that tellsuwotto return the UMAP model so that we can transform new data later.
mnist_train_umap <-
umap(
mnist_train,
nn_method = "hnsw",
batch = TRUE,
n_epochs = 500,
n_sgd_threads = 6,
ret_model = TRUE,
verbose = TRUE
)UMAP embedding parameters a = 1.896 b = 0.8006
Converting dataframe to numerical matrix
Read 60000 rows and found 784 numeric columns
Building HNSW index with metric 'l2' ef = 200 M = 16 using 6 threads
Finished building index
Searching HNSW index with ef = 15 and 6 threads
Finished searching
Commencing smooth kNN distance calibration using 6 threads with target n_neighbors = 15
Initializing from normalized Laplacian + noise (using RSpectra)
Commencing optimization for 500 epochs, with 1239066 positive edges using 6 threads
Using method 'umap'
Optimizing with Adam alpha = 1 beta1 = 0.5 beta2 = 0.9 eps = 1e-07
0% 10 20 30 40 50 60 70 80 90 100%
[----|----|----|----|----|----|----|----|----|----|
**************************************************|
Optimization finishedUMAP on test data
You don’t need to specify the use of HNSW for transforming new data,
the model contains the necessary information. So we can just use the
umap_transform as normal:
mnist_test_umap <-
umap_transform(
X = mnist_test,
model = mnist_train_umap,
n_sgd_threads = 6,
verbose = TRUE
)Read 10000 rows and found 784 numeric columns
Processing block 1 of 1
Finished searching
Commencing smooth kNN distance calibration using 6 threads with target n_neighbors = 15
Initializing by weighted average of neighbor coordinates using 6 threads
Commencing optimization for 167 epochs, with 150000 positive edges using 6 threads
Using method 'umap'
Optimizing with Adam alpha = 1 beta1 = 0.5 beta2 = 0.9 eps = 1e-07
0% 10 20 30 40 50 60 70 80 90 100%
[----|----|----|----|----|----|----|----|----|----|
**************************************************|
FinishedPlotting the results
Let’s plot the embeddings to make sure that HNSW is giving us useful
results. I will use ggplot2, and the
Polychrome package to create a set of distinct colors for
each digit.
install.packages(c("ggplot2", "Polychrome"))
library(ggplot2)
library(Polychrome)This gives a palette of 10 colors following a recipe that is a bit like that in the Python glasbey package:
palette <- as.vector(Polychrome::createPalette(
length(levels(mnist$Label)) + 2,
seedcolors = c("#ffffff", "#000000"),
range = c(10, 90)
)[-(1:2)])Now we can plot the training and test data, which I will do in two separate plots.
ggplot(
data.frame(mnist_train_umap$embedding, Digit = mnist_train$Label),
aes(x = X1, y = X2, color = Digit)
) +
geom_point(alpha = 0.5, size = 1.0) +
scale_color_manual(values = palette) +
theme_minimal() +
labs(
title = "MNIST training set UMAP",
x = "",
y = "",
color = "Digit"
) +
theme(legend.position = "right") +
guides(color = guide_legend(override.aes = list(size = 5, alpha = 1)))
ggplot(
data.frame(mnist_test_umap, Digit = mnist_test$Label),
aes(x = X1, y = X2, color = Digit)
) +
geom_point(alpha = 0.5, size = 1.0) +
scale_color_manual(values = palette) +
theme_minimal() +
labs(
title = "MNIST test set UMAP",
x = "",
y = "",
color = "Digit"
) +
theme(legend.position = "right") +
guides(color = guide_legend(override.aes = list(size = 5, alpha = 1)))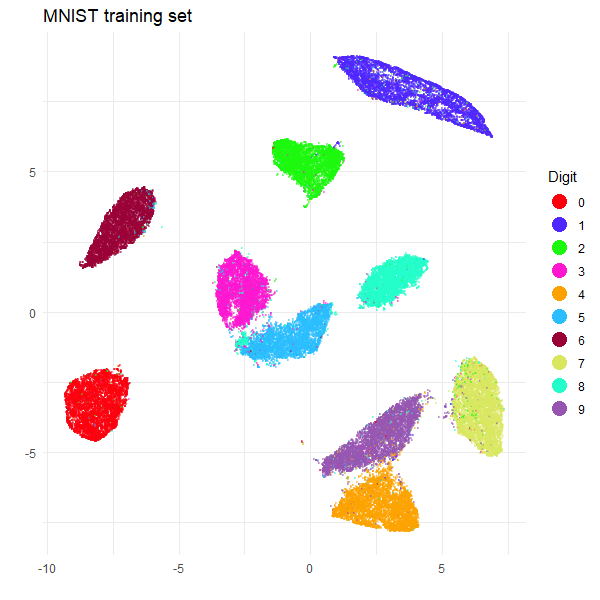
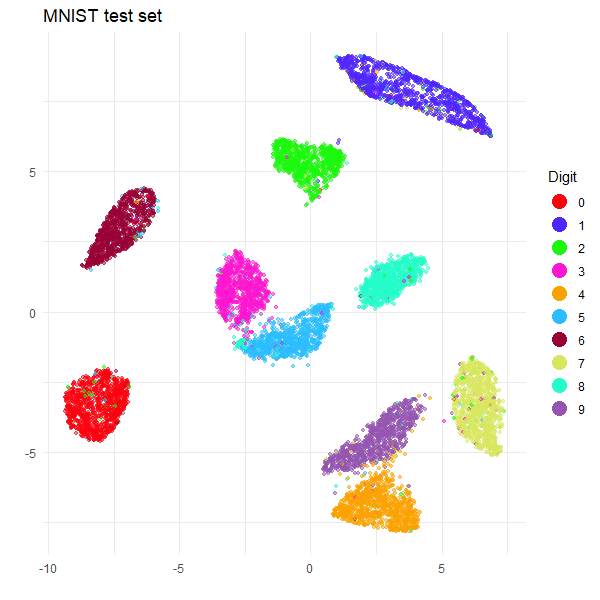
The training set embedding looks like a very typical UMAP-on-MNIST result, and the test clusters are all in the right place relative to the training set. So we can celebrate victory and a successful use of HNSW.
HNSW parameters
umap has a number of parameters that can be set to
control the behavior of nearest neighbor search with its default method,
Annoy, namely n_trees and search_k. These are
not used when nn_method = "hnsw" is set. Instead, you can
control the behavior of the HNSW index by passing a list of arguments as
nn_args. For HNSW, you can set:
-
M: The number of bi-directional links created for every new element during construction. Higher values mean more accurate search, but slower construction and larger index size. Default is 16. -
ef_construction: The size of the dynamic list for the nearest neighbors during the construction. Higher values mean more accurate search, but slower construction and larger index size. Default is 200. -
ef: The size of the dynamic list for the nearest neighbors during the search. Higher values mean more accurate search, but slower search. Default is 10, but it will be set ton_neighborsif the latter is larger.
For example you could do something like:
Using RcppHNSW Directly
The nn_method = "hnsw" is a new feature in
uwot. In previous versions, you could still make use of
HNSW neighbors but you would need to work with RcppHNSW
directly. The rest of this article shows you how to do this, and can be
used as a way to understand how RcppHNSW works and how to
use external nearest neighbor data with uwot.
Preparing the data
Our starting point is still mnist_train and
mnist_test. As a reminder, these are dataframes with both
the numerical pixel intensities of the images as well as the
Label factor describing the number the digit
represents.
We won’t use the labels for calculating nearest neighbors, and in
fact RcppHNSW is a bit more fussy than uwot.
It only wants numerical matrix data only for input, so we will define a
function to convert the dataframe to a matrix containing only the
numerical data and then generate the matrices we need.
Using RcppHNSW
Now load RcppHNSW:
Time to build an index using the training data. The default settings
for hnsw_build are perfectly good for MNIST, so we’ll go
with that. I recommend using as many threads as you can for this stage.
Here I use 6 threads:
mnist_index <- hnsw_build(mnist_train_data, n_threads = 6)MNIST training data k-nearest neighbors
Now we will query the index with the training data to find the k-nearest neighbors of the training data. Note that in this case we built the index with the same data that we are querying it with.
As with the index building, we can get good performance with default
settings, but it’s recommended to use as many threads as you can. The
searching should be substantially faster than the index building,
though. The only other extra parameter we need is to specify the number
of neighbors we want. In uwot, the default number of
neighbors is 15, so we shall use that for the k
parameter:
mnist_train_knn <-
hnsw_search(
mnist_train_data,
mnist_index,
k = 15,
n_threads = 6
)There is a separate article
on uwot’s nearest neighbor format but the good news is
that the output format from RcppHNSW is already in the right format for
uwot (this is not a coincidence: I created and maintain the
RcppHNSW package). Nonetheless, let’s take a look at the output, which
is a list of two matrices:
names(mnist_train_knn)The first matrix, idx, contains the indices of the
nearest neighbors for each point in the training data. The second
matrix, dist, contains the distances to the nearest
neighbors. Let’s take a look at the dimensions of these matrices:
lapply(mnist_train_knn, `dim`)So we have 60,000 rows, one for each point in the training data, and 15 columns, one for each nearest neighbor. Let’s take a look at the first few rows and columns of each matrix:
mnist_train_knn$idx[1:3, 1:3]
mnist_train_knn$dist[1:3, 1:3]For the k-nearest neighbors of a dataset, it’s often a convention
that the nearest neighbor of any item is the item itself, which is why
the nearest neighbor of the first item has the index 1, and
the distance is zero. Not all nearest neighbor packages follow this
convention, but uwot does, so we’re good.
MNIST test set query neighbors
We will also need the test set neighbors, so let’s do that now. We are not going to build an index with the test set, but instead we will query each test set item against the training set index:
mnist_test_query_neighbors <-
hnsw_search(
mnist_test_data,
mnist_index,
k = 15,
n_threads = 6,
verbose = TRUE
)The output is the same format as for the training set neighbors, so there should be no surprises here:
lapply(mnist_test_query_neighbors, `dim`)Here are the first few indices and distances for the test set:
mnist_test_query_neighbors$idx[1:3, 1:3]
mnist_test_query_neighbors$dist[1:3, 1:3] [,1] [,2] [,3]
[1,] 676.5841 793.9868 862.6766
[2,] 1162.9316 1211.8445 1285.9285
[3,] 321.6629 332.4635 341.0484Remember that we queried the test set against the training set, so the test set indices don’t get a chance to be neighbors of themselves. Consequently, also the nearest neighbor distances are not zero.
Using HNSW k-nearest neighbors with UMAP
We are now ready to use the HNSW k-nearest neighbors with
uwot.
UMAP on training data with HNSW knn
UMAP works on the nearest neighbor graph, so if you pass it pre-computed nearest neighbor data, you don’t actually need to give it any other data. To do so:
- set
X = NULL. - pass the nearest neighbor data as the
nn_methodparameter.
The other parameters are the same as those we used before with
nn_method = "hnsw", so see above for a description.
mnist_train_umap <-
umap(
X = NULL,
nn_method = mnist_train_knn,
batch = TRUE,
n_epochs = 500,
n_sgd_threads = 6,
ret_model = TRUE,
verbose = TRUE
)UMAP embedding parameters a = 1.896 b = 0.8006
Commencing smooth kNN distance calibration using 6 threads with target n_neighbors = 15
Initializing from normalized Laplacian + noise (using irlba)
Commencing optimization for 500 epochs, with 1239236 positive edges using 6 threads
Using method 'umap'
Optimizing with Adam alpha = 1 beta1 = 0.5 beta2 = 0.9 eps = 1e-07
0% 10 20 30 40 50 60 70 80 90 100%
[----|----|----|----|----|----|----|----|----|----|
**************************************************|
Optimization finished
Note: model requested with precomputed neighbors. For transforming new data, distance data must be provided separatelyNothing very exciting is in the output, except that notice the last line of output reminding us that if we want to transform new data we better have generated neighbors for that data too. And we did, so we’re ok.
Transforming the test data
Let us now transform the test data using the UMAP model we just
created. There are far fewer nobs to twiddle when transforming new data
as that is mainly baked into the UMAP model. Again, we pass
X = NULL as we don’t need the original test set data, and
pass the test set nearest neighbor data as the nn_method
parameter. We also need to pass the UMAP model we just created.
mnist_test_umap <-
umap_transform(
X = NULL,
model = mnist_train_umap,
nn_method = mnist_test_query_neighbors,
n_sgd_threads = 6,
verbose = TRUE
)Read 10000 rows
Processing block 1 of 1
Commencing smooth kNN distance calibration using 6 threads with target n_neighbors = 15
Initializing by weighted average of neighbor coordinates using 6 threads
Commencing optimization for 167 epochs, with 150000 positive edges using 6 threads
Using method 'umap'
Optimizing with Adam alpha = 1 beta1 = 0.5 beta2 = 0.9 eps = 1e-07
0% 10 20 30 40 50 60 70 80 90 100%
[----|----|----|----|----|----|----|----|----|----|
**************************************************|
FinishedAt this point we could visualize the data again, but you can take my
word for it that it looks just like the nn_method = "hnsw"
approach.
Conclusions
To summarize, to use HNSW with uwot do the
following:
- Make sure
RcppHNSWis installed. - Set
nn_method = "hsnw". - Pass an
nn_argslist containing a combination ofM,ef_constructionandefto control the speed/accuracy trade-off. - To transform new data, just use
umap_transformas normal.
To use RcppHNSW directly for UMAP do the following:
- build an index for your data with
hnsw_build. - query the index with
hnsw_searchto obtain the k-nearest neighbors. - run UMAP on it with
umapsetting passing the neighbor data tonn_method.
To transform new data, do the same thing with the following additions and modifications:
- use
hnsw_searchwith the new data and the index created in the first step above to get the neighbors for your new data (with respect to the training data). - when you run UMAP, set
ret_model = TRUEto get the UMAP model back. - use
umap_transformwith the UMAP model and pass the new data’s neighbors asnn_method.
You also don’t need to keep the original data around once you have
the neighbors, so you can set X = NULL when running
umap and umap_transform. But that data can
still be useful for initialization. For example if you want to use a
PCA-based initialization then you will need to keep your
original data around. But it’s not necessary with the default settings
for umap.